
Frequently Asked Questions
General
-
-
non-B DB is a database that stores candidate non-B DNA structures that are systematically identified from the genomic regions of several mammalian species. non-B DB also allows researchers to intersect non-B DNA information with positions of known variation in the genome to assess the possible disruption of these structures as a function of genotypic variation.
-
-
-
Several recent publications have provided significant evidence that non-B DNA structures may play a role in DNA instability and mutagenesis, leading to both DNA rearrangements and increased mutational rates, which are hallmark of cancer. The goal of non-B DB is to accelerate cancer research by helping cancer researchers find therapeutic treatments for several cancer types including but not limited to breast, ovary, glioblastoma, prostrate, etc.
-
-
-
DNA exists in many possible conformations that include the A-DNA, B-DNA, and Z-DNA forms; of these, B-DNA is the most common form found in cells. The DNAs that do not fall into a right-handed Watson-Crick double-helix are known as non-B DNAs and comprise cruciform, triplex, slipped (hairpin) structures, tetraplex (G-quadruplex), left-handed Z-DNA, and others.
-
-
-
Currently, non-B DB provides the most complete list of alternative DNA motif predictions available, including Z-DNA motifs, quadruplex forming motifs, inverted repeats, mirror repeats and direct repeats and their associated subsets of cruciforms, triplex and slipped structures, respectively. The database also contains motifs predicted to form static DNA bends, short tandem repeats and homo(purine.pyrimidine) tracts.
-
-
-
Since non-B DB contains information on all the main forms of non-B DNA, it is the most comprehensive database of its kind. Other existing databases cover one type of non-B DNA. In addition to the wider scope of coverage, non-B DB was built upon the most updated genome assembly available as of today (e.g. hg19 (build 38) for human). In addition, non-B DB enables users to perform different types of query and visualize the results using GBrowse 2.0 which allows multiple data sources.
-
-
-
We have taken the approach of using rather broad and general identification methods based exclusively on sequence features; thus, although subsequent filtering of the sampled data is straightforward because of the flexibility provided by the database, our current criteria are expected to include a subset of both false positive and negative hits.
Input from the community regarding enhanced algorithms for the detection or scoring of identified motifs is most welcomed and may be incorporated into the system if appropriate. Please feel free to contact us.DNA feature Search Criteria Subset of "DNA feature" forming non-B DNA Search criteria for "Subset of DNA feature" Example Inverted Repeat 10–100 nt with reverse complement within 100 nt spacer Cruciform_Motif if spacer=0-3 nt 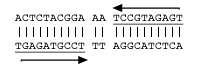
Mirror Repeat 10–100 nt mirrored within 100 nt spacer Triplex_Motif 90% Purine or Pyrimidine and 0–8 nt spacer 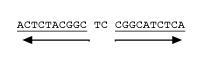
Direct Repeat 10–50 nt repeated within 5 nt spacer Slipped_Motif if spacer=0 nt 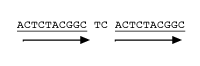
G-Quadruplex Forming Repeats 4 or more G-tracts (3-7 G’s) separated by 1–7 nt spacers; Preference for short spacers with C’s and/or T’s Whole set As per the whole set 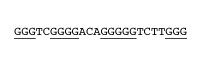
Z-DNA Repeat G followed by Y (C or T) for at least 10 nt; One strand must be alternating Gs Whole set As per the whole set 
A-Phased Repeats 3 or more A-tracts (3-5 As) 10 nt on center each; Spacers between equal sized A-tracts must contain some non As Whole set As per the whole set 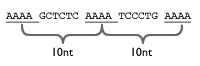
-
-
-
Acronym Description A Adenine ABCC Advanced Biomedical Computing Center APR A-Phased Repeat C Cytosine CMP composition DAS Distributed Annotation System DR Direct Repeat G Guanine GFF General Feature Format GQFR G-Quadruplex Forming Repeat IR Inverted Repeat MB Megabyte MR Mirror Repeat nBMST non-B DNA Motif Search Tool R Purine rsrd right strand right direction (in reference to DNA strand orientation) STR Short Tandem Repeat T Thymine wswd wrong strand wrong direction (in reference to DNA strand orientation) Y Pyrimidine ZDM Z-DNA Motif
-
-
-
Please visit the registration page .
-
Genomic Database Search Tools
-
-
Search by Feature allows you to search the non-B DB by specifying one or more genomic features.
Search by Location allows you to get location-specific annotations. Enter chromosome, start, and stop information and all the genomic features found within the region will be returned in a tabular format.
-
-
-
DAS stands for Distributed Sequence Annotation System. The four DAS available on the non-B DB are as follows:
DAS Description abccDAS This database includes features computed at the ABCC including PuPys, STRs, composition, physical DNA characteristics, gene based synteny blocks (GBSB), etc. mappingDAS This database includes features remapped to the reference genome using gmap for other data sources such as RefSeq, Ensembl, MGC, Unigene,miRBase etc. ncbiDAS This database includes features derived from the NCBI genomes directories including genes, SNPs, cytogenic markers assembly information, RepeatMasker elements, etc. nonbDAS This database ncludes alternative DNA structure predictions, including Z-DNA motifs, g-quadruplex forming motifs, inverted repeats, mirror repeats and direct repeats and their associated subsets of cruciforms, triplex and slipped structures, respectively.
-
-
-
A Phased Repeat
- Composition (Equals, Not Equal To)
- Text string signifying the number of A, C, G, and T nucleotides in the motif sequence. Values of 0 or greater are allowed for each nucleotide.
- Example=15A/3C/2G/6T
- Sequence (Equals, Not Equal To)
- Text string signifying the exact motif pattern String may consist of lowercase 'a', 't', 'c', and 'g' characters without spaces.
- Example=aaaaattttcaacaaaatactaggaaa
- Tracts (Equals, Not Equal To, Greater Than, Less Than)
- Integer value of 3 or greater signifying the number of consecutive adenine tracts making up the A Phased Repeat.
- Example=3
Direct Repeat
- Spacer(Equals, Not Equal To, Greater Than, Less Than)
- Integer value of 0 or greater signifying the number of nucleotides in the spacer region of the motif.
- Example=2
- Repeat (Equals, Not Equal To, Greater Than, Less Than)
- Integer value of 10 or greater signifying the number of nucleotides in the repeated portion of motif.
- Example=10
- Composition (Equals, Not Equal To)
- Text string signifying the number of A, C, G, and T nucleotides in the repeat portion of the motif sequence. Values of 0 or greater are allowed for each nucleotide.
- Example=8A/2C/1G/5T
- Sequence (Equals, Not Equal To)
- Text string signifying the exact motif nucleotide sequence. String may consist of lowercase “aâ€, tâ€, “câ€, and “g†characters without spaces.
- Example=ttccaaaataattgaataattccaaaataattgaa
- Subset (Equals, Not Equal To)
- Integer value or 0 or 1 signifying if the motif meets the requirements for identification as a subset of the main motif type. Motifs with “subset=1†meet this more restrictive criteria and are thought to be likely to be able to form non-B DNA structures in vivo.
- Example=1
G Quadruplex Motif
- Composition (Equals, Not Equal To)
- Text string signifying the number of A, C, G, and T nucleotides in the motif sequence. Values of 0 or greater are allowed for each nucleotide.
- Example=6A/1C/18G/1T
- Sequence (Equals, Not Equal To)
- Text string signifying the exact motif nucleotide sequence. String may consist of lowercase “aâ€, tâ€, “câ€, and “g†characters without spaces.
- Example=gggaggagggaggaaggggggcatggg
- Islands (Equals, Not Equal To, Greater Than, Less Than)
- Integer value of 1 or greater signifying the number of uninterrupted stretches of guanines that make up the G Quadruplex motif.
- Example=4
- Runs (Equals, Not Equal To, Greater Than, Less Than)
- Integer value of 4 or greater signifying the number of possible non-adjacent runs of 3 or more guanines making up the G Quadruplex motif.
- Example=4
- Max (Equals, Not Equal To, Greater Than, Less Than)
- Integer value of 3 or larger signifying the maximum guanine run length for which 4 or more consecutive runs can be formed in the motif.
- Example=3
Inverted Repeat
- Spacer(Equals, Not Equal To, Greater Than, Less Than)
- Integer value of 0 or greater signifying the number of nucleotides in the spacer region of the motif.
- Example=2
- Repeat (Equals, Not Equal To, Greater Than, Less Than)
- Integer value of 6 or greater signifying the number of nucleotides in the repeated portion of motif.
- Example=6
- Perms (Equals, Not Equal To, Greater Than, Less Than)
- Integer value of 1 or greater signifying the number of times one of half of the repeated sequence can be shifted to create multiple matches with the other half.
- Example=1
- Minloop (Not relevant, should be removed as an option)
- Composition (Equals, Not Equal To)
- Text string signifying the number of A, C, G, and T nucleotides in the repeat portion of the motif sequence. Values of 0 or greater are allowed for each nucleotide.
- Example=0A/1C/0G/5T
- Sequence (Equals, Not Equal To)
- Text string signifying the exact motif nucleotide sequence. String may consist of lowercase “aâ€, tâ€, “câ€, and “g†characters without spaces.
- Example=ttctttaaaaagaa
- Subset (Equals, Not Equal To)
- Integer value or 0 or 1 signifying if the motif meets the requirements for identification as a subset of the main motif type. Motifs with “subset=1†meet this more restrictive criteria and are thought to be likely to be able to form non-B DNA structures in vivo.
- Example=1
Mirror Repeat
- Spacer(Equals, Not Equal To, Greater Than, Less Than)
- Integer value of 0 or greater signifying the number of nucleotides in the spacer region of the motif.
- Example=2
- Repeat (Equals, Not Equal To, Greater Than, Less Than)
- Integer value of 10 or greater signifying the number of nucleotides in the repeated portion of motif.
- Example=10
- Perms (Equals, Not Equal To, Greater Than, Less Than)
- Integer value of 1 or greater signifying the number of times one of half of the repeated sequence can be shifted to create multiple matches with the other half.
- Example=1
- Minloop (Not relevant, should be removed as an option)
- Composition (Equals, Not Equal To)
- Text string signifying the number of A, C, G, and T nucleotides in the repeat portion of the motif sequence. Values of 0 or greater are allowed for each nucleotide.
- Example=0A/7C/0G/3T
- Sequence (Equals, Not Equal To)
- Text string signifying the exact motif nucleotide sequence. String may consist of lowercase “aâ€, tâ€, “câ€, and “g†characters without spaces.
- Example=cctcctcctcggcatactcctcatcctcctcctcc
- Subset (Equals, Not Equal To)
- Integer value or 0 or 1 signifying if the motif meets the requirements for identification as a subset of the main motif type. Motifs with “subset=1†meet this more restrictive criteria and are thought to be likely to be able to form non-B DNA structures in vivo.
- Example=1
Short Tandem Repeat
- Composition (Equals, Not Equal To)
- Text string signifying the number of A, C, G, and T nucleotides in the motif sequence. Values of 0 or greater are allowed for each nucleotide.
- Example=1A/0C/2G/0T
- Sequence (Equals, Not Equal To)
- Text string signifying the exact motif pattern String may consist of lowercase “aâ€, tâ€, “câ€, and “g†characters without spaces.
- Example=ggaggaggag
- Length (Equals, Not Equal To, Greater Than, Less Than)
- Integer value from 1 to 9 signifying the length of the short repeated sequence.
- Exampe=1
- Type(Equals, Not Equal To, Greater Than, Less Than)
- Integer value from 1 to 11representing the properties of odd, RY/YR pattern, symmetric, and complementary sequences encoded by subsequent binary bits for the short repeated sequence. For example, a value of 4 signifies an odd length symmetric repeated sequence (e.g. “ctcâ€) .
- Exampe=1
Z DNA Motif
- Composition (Equals, Not Equal To)
- Text string signifying the number of A, C, G, and T nucleotides in the motif sequence. Values of 0 or greater are allowed for each nucleotide.
- Example=2A/3C/3G/3T
- Sequence (Equals, Not Equal To)
- Text string signifying the exact motif pattern String may consist of lowercase “aâ€, tâ€, “câ€, and “g†characters without spaces.
- Example=tgtgtgcacaca
- Subset (being removed from current version)
- Length (Equals, Not Equal To, Greater Than, Less Than)
- Integer 10 or greater signifying the length of the Z-DNA motif.
- Example=10
- Score (being removed from current version)
-
Miscellaneous
-
-
There are two ways to contact us:(1) At the top of this FAQ page, click on Submit a Question.(2) On the side menu, click on Contact us.
-
non-B Motif Search Tool (nBMST)
-
-
Yes. On the nBMST page, there are two example sequences for demonstration purpose: a single FASTA sequence and multi FASTA sequences.
-
-
-
In the GFF output file, the last column contains the actual sequence information.For example,chr8 ABCC G-Quadruplex_Forming_Repeat 1603 1630 . + . ID=chr8_1603_1630_GQFR_rsrd;Length=28;BestStruct=G4-N7-G4-N2-G4-N2-G4(CGGTACT/GT/AC);NbrStructs=8;Sequence=GGGGCGGTACTGGGGGTGGGGACGGGGG;BestScore=16;
-
-
 Can I specify how long my repeats can be, the length of the spacer(loops) and the inclusion of mismatches in this nBMST?
Can I specify how long my repeats can be, the length of the spacer(loops) and the inclusion of mismatches in this nBMST?
-
 Why do I see two types of motifs in one selection? For instance, mirror repeats and triplex motifs. Why can't I select only one type of motif (i.e. only mirror repeats)?
Why do I see two types of motifs in one selection? For instance, mirror repeats and triplex motifs. Why can't I select only one type of motif (i.e. only mirror repeats)?
-
Our computational algorithms search for patterns matching a certain type of non-B DNA and its subset. Therefore, when you search for mirror repeats, you will also be given the subset of triplex forming motifs. The same applies for inverted repeats and the subset of cruciform motifs, and direct repeats and the subset of slipped motifs.
-
-
-
Yes, we allow up to 20 MB of data to be uploaded. If you need to analyze a larger file size, please feel free to contact us. We will be happy to assist you.
-
-
-
Turnaround time for results will vary depending on the size of the sequence(s), the number and types of the non-B DNA motifs selected, and the computing resources available at the ABCC at the time of submission.In cases where input sequences are very large and/or multiple motifs are selected, an email address is recommended to avoid waiting on-line. A notification email will be sent when the job is completed.It should be noted that the algorithms for mirror and inverted repeats are the most computationally intensive, and therefore take more time to complete than the rest of the motifs. Thus, in cases where quick results are desired for large sequences, it is recommended that separate jobs be submitted for these two motif types.
-
non-B DB v2.0
-
-
(1) We have further enhanced search criteria significantly and optimized the algorithms and re-annotated all the five species, human, mouse, chimp, macaque, and dog from v1.0.
(2) We have 7 additional species since v1.0 namely arabidopsis thaliana, cow, horse, orangutan, pig, platypus, and rat.
(3) We have included extensive visualization plots using whole genomic level Circos plots and detailed chromosomal level R plots in addition to the existing PolyBrowse viewer.
(4) We have consolidated Search by Feature and Search by Feature Attributes.
-